Analysis-Random-Forest.r
Mushroom Data Source archive.ics.uci.edu
Load the needed R libraries
#Load needed librarys
source('prepare_functions.R')
library(randomForest)
library(e1071)
library(caret)
library(ggplot2)
#Set a consistent seed
set.seed(123)
#Major clean and preparation see helper_function.r
data = prepareAndCleanData()
#Check the first couple of rows of data
head(data)
Just a few of the 23 variables
Edible CapShape CapSurface CapColor Bruises Odor GillAttachment
1 Poisonous Convex Smooth Brown True Pungent Free
2 Edible Convex Smooth Yellow True Almond Free
3 Edible Bell Smooth White True Anise Free
4 Poisonous Convex Scaly White True Pungent Free
5 Edible Convex Smooth Gray False None Free
6 Edible Convex Scaly Yellow True Almond Free
Create data for training and testing.
This is the back bone for machine learning. We are taking the data and splitting into two groups one is for the model the other is to run through the model, to train the model. The test data will be ~7700 records and the training data will be around ~400 records.
sample.ind = sample(2,
nrow(data),
replace = T,
prob = c(0.05,0.95))
data.dev = data[sample.ind==1,]
data.val = data[sample.ind==2,]
The data sets look good as edible vs poisonous proportion ratio is about equal To see if the traning and test data are some what equal
table(data$Edible)/nrow(data)
Edible Poisonous
0.5179714 0.4820286
table(data.dev$Edible)/nrow(data.dev)
Edible Poisonous
0.5025 0.4975
table(data.val$Edible)/nrow(data.val)
Edible Poisonous
0.5187727 0.4812273
Lets run the random forest algorithm with number of trees being 100 The more the better, but we don’t want to over do it.
rf = randomForest(Edible ~ .,
ntree = 100,
data = data.dev)
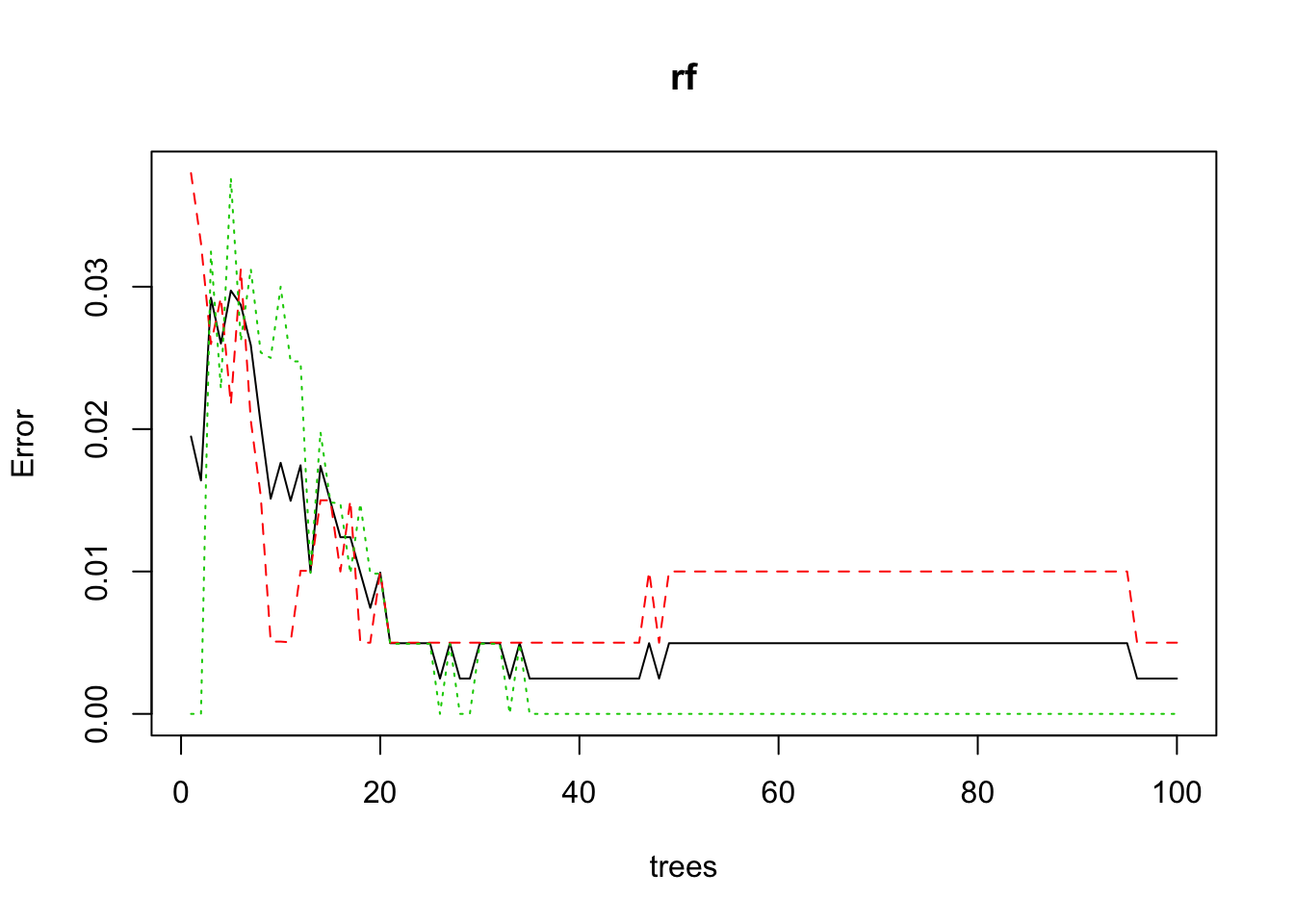
plot(rf)
rf plot
This plot will tell us if the number of trees we picked will give good results. As you can see 10-30 trees causes a minor error span.
Plot the top 10 variables for prediction of edible vs posionous.
varImpPlot(rf,
sort = T,
n.var=10,
main="Top 10 - Variable Predictors")
Top 10 Predictors
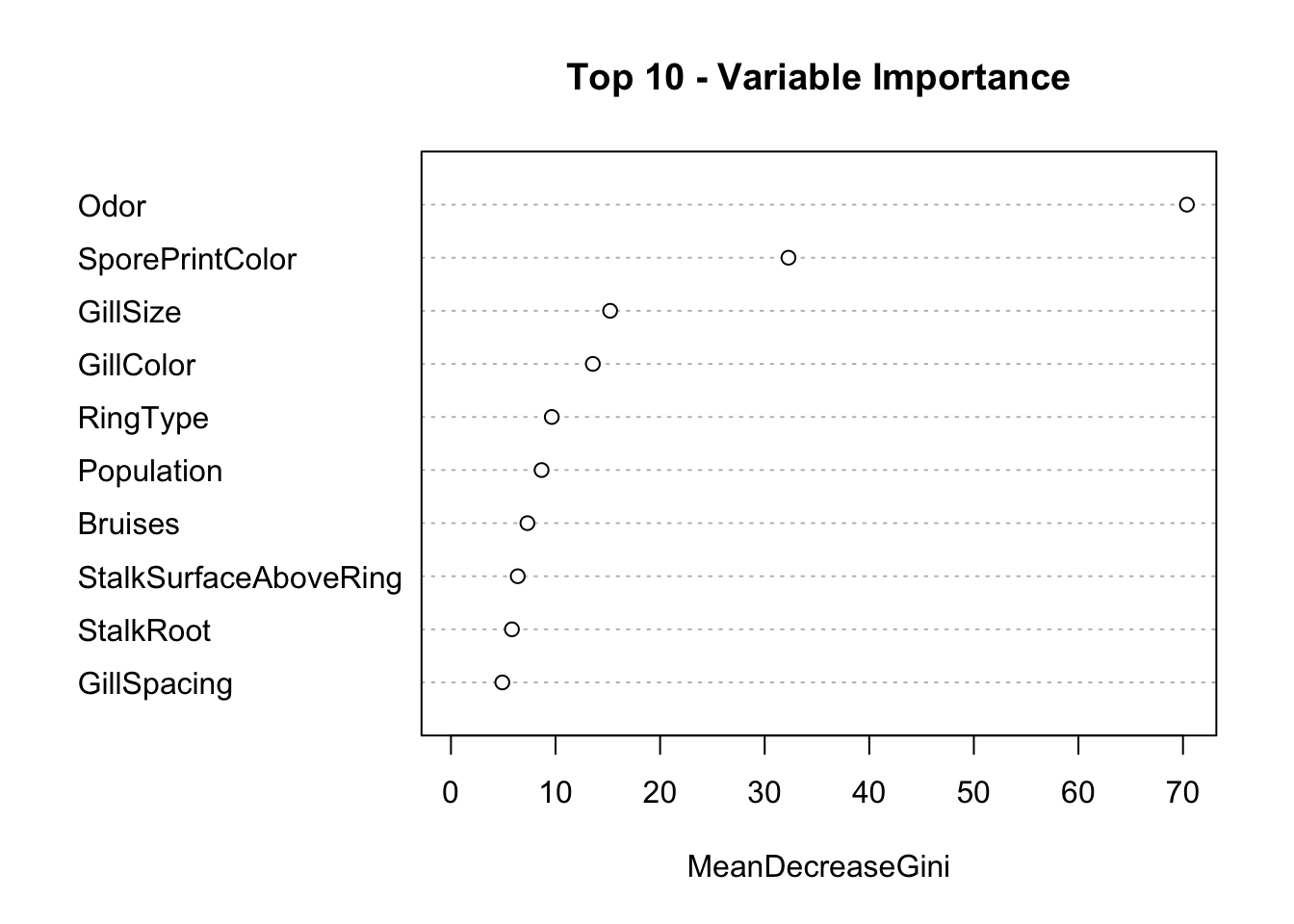
Looks like Odor is the greatest indicator along with Spore Print Color
var.imp = data.frame(importance(rf,
type=2))
#Make row names as columns
var.imp$Variables <- row.names(var.imp)
var.imp[order(var.imp$MeanDecreaseGini,decreasing = T),]
MeanDecreaseGini Variables
Odor 70.62464550 Odor
SporePrintColor 34.98215873 SporePrintColor
GillColor 14.38042458 GillColor
RingType 11.25159749 RingType
GillSize 11.12214770 GillSize
Population 8.39424526 Population
StalkSurfaceAboveRing 6.91877682 StalkSurfaceAboveRing
Habitat 6.53783046 Habitat
StalkRoot 5.61593844 StalkRoot
StalkSurfaceBelowRing 5.59337397 StalkSurfaceBelowRing
Bruises 5.47550779 Bruises
StalkColorBelowRing 5.08844477 StalkColorBelowRing
CapColor 3.29438185 CapColor
GillSpacing 2.74912949 GillSpacing
StalkColorAboveRing 2.26185667 StalkColorAboveRing
StalkShape 1.79067923 StalkShape
CapSurface 1.10385529 CapSurface
RingNumber 0.93590714 RingNumber
CapShape 0.61875375 CapShape
VeilColor 0.13948555 VeilColor
GillAttachment 0.01928571 GillAttachment
VeilType 0.00000000 VeilType
Here is the 400 samples ran through the model the confidence level is 99%, meaning we can say that we can predict mushroom edibility 99% of the time. See the Confusion Matrix and Statistics below.
#Predicting response variable
data.dev$predicted.response <- predict(rf , data.dev)
# Create Confusion Matrix for the traning data
#Calculates a cross-tabulation of observed and predicted classes with associated statistics.
confusionMatrix(data = data.dev$predicted.response,
reference = data.dev$Edible,
positive = 'Edible')
## Confusion Matrix and Statistics
##
## Reference
## Prediction Edible Poisonous
## Edible 201 0
## Poisonous 0 199
##
## Accuracy : 1
## 95% CI : (0.9908, 1)
## No Information Rate : 0.5025
## P-Value [Acc > NIR] : < 2.2e-16
##
## Kappa : 1
## Mcnemar's Test P-Value : NA
##
## Sensitivity : 1.0000
## Specificity : 1.0000
## Pos Pred Value : 1.0000
## Neg Pred Value : 1.0000
## Prevalence : 0.5025
## Detection Rate : 0.5025
## Detection Prevalence : 0.5025
## Balanced Accuracy : 1.0000
##
## 'Positive' Class : Edible
##
#Predicting response variable
data.val$predicted.response <- predict(rf ,data.val)
#Create Confusion Matrix for the test data
#Calculates a cross-tabulation of observed and predicted classes with associated statistics.
confusionMatrix(data=data.val$predicted.response,
reference=data.val$Edible,
positive='Edible')
## Confusion Matrix and Statistics
##
## Reference
## Prediction Edible Poisonous
## Edible 4007 8
## Poisonous 0 3709
##
## Accuracy : 0.999
## 95% CI : (0.998, 0.9996)
## No Information Rate : 0.5188
## P-Value [Acc > NIR] : < 2e-16
##
## Kappa : 0.9979
## Mcnemar's Test P-Value : 0.01333
##
## Sensitivity : 1.0000
## Specificity : 0.9978
## Pos Pred Value : 0.9980
## Neg Pred Value : 1.0000
## Prevalence : 0.5188
## Detection Rate : 0.5188
## Detection Prevalence : 0.5198
## Balanced Accuracy : 0.9989
##
## 'Positive' Class : Edible
##
With odor and spore print color we can predict almost 99% of the time.
#Lets plot some jitter classification plots to show what odors and spore print colors show the poisonous mushrooms
#This plot shows that CapShape and CapSurface really dont give a good indication of edible mushrooms
p = ggplot(data,aes(x=CapShape, y=CapSurface, color=Edible))
p + geom_jitter(alpha=0.3) + scale_color_manual(breaks = c('Edible','Poisonous'),values=c('blue','orange'))
ggPlot Jitter Plots
CapShape vs CapSurface 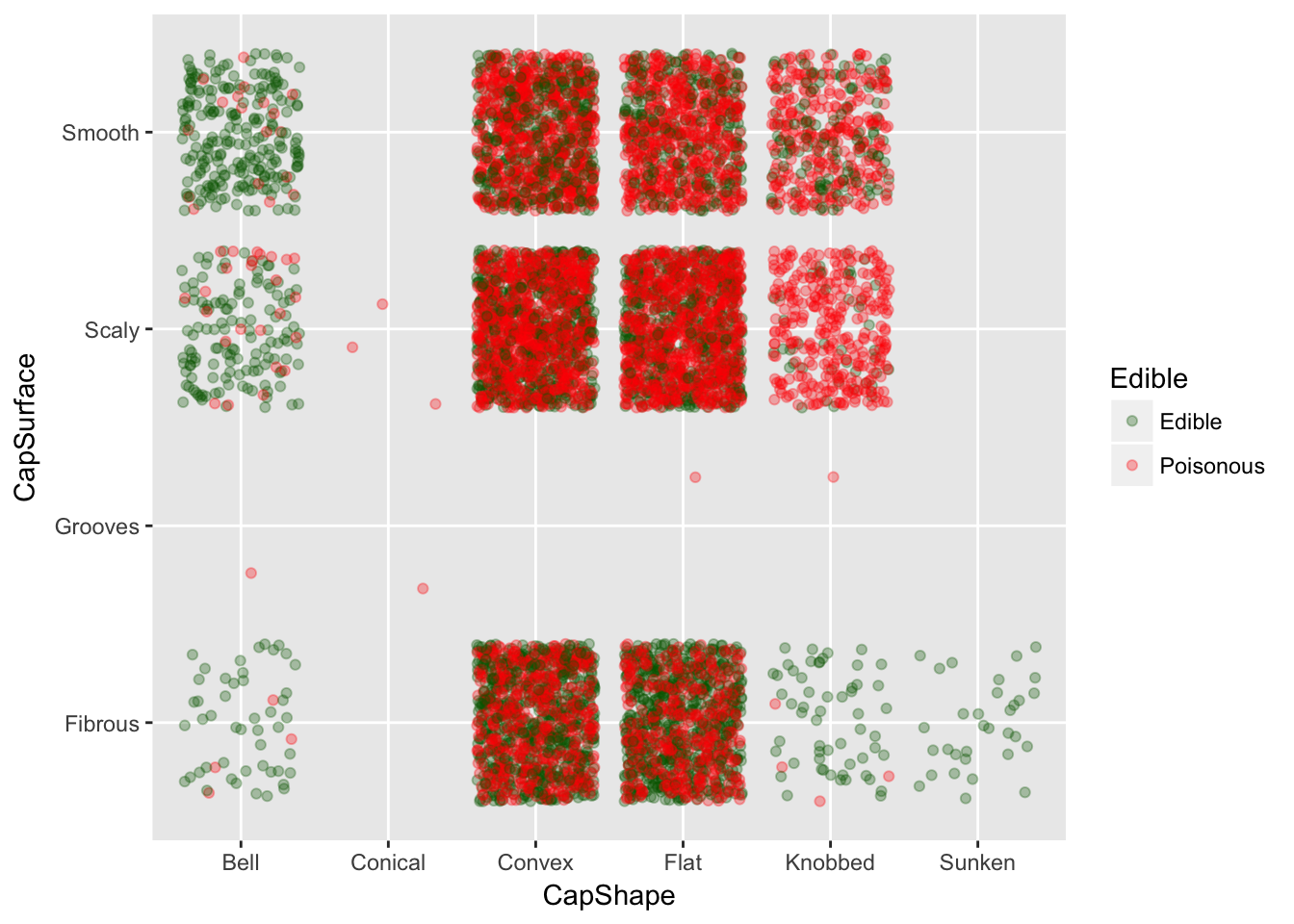
Odor vs Spore Print Color 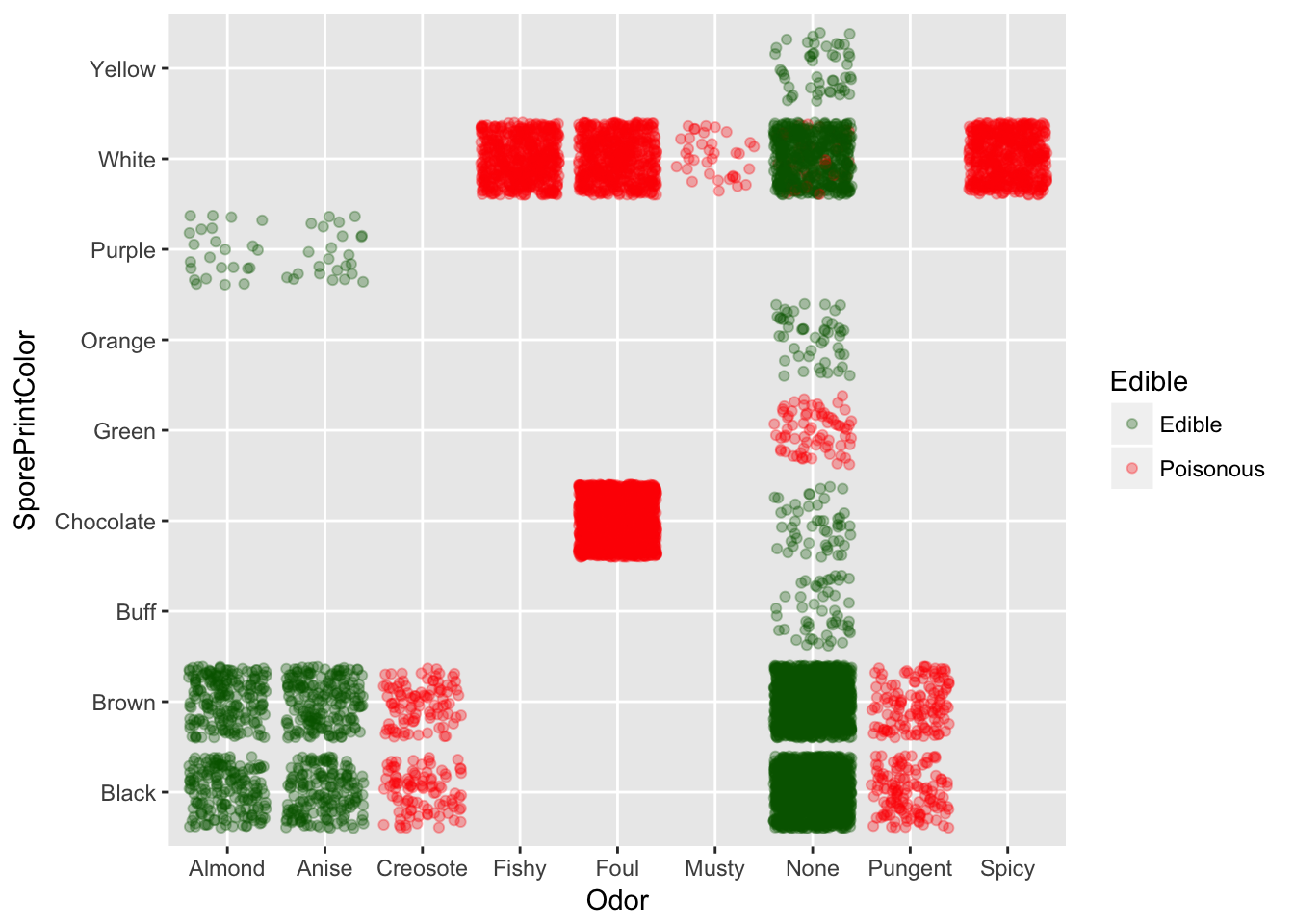
Odor vs Edible 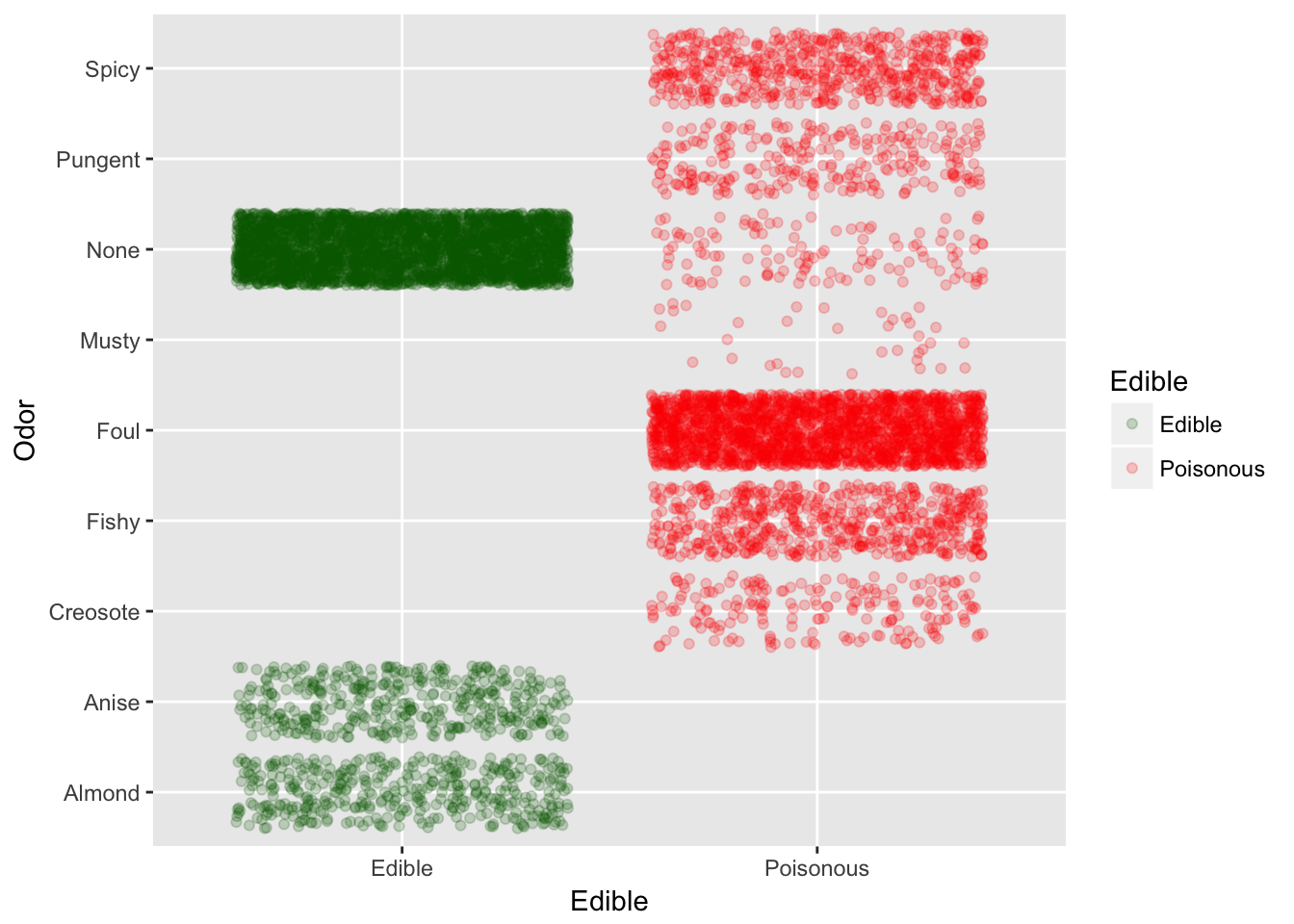
Spore Print Color vs Edible 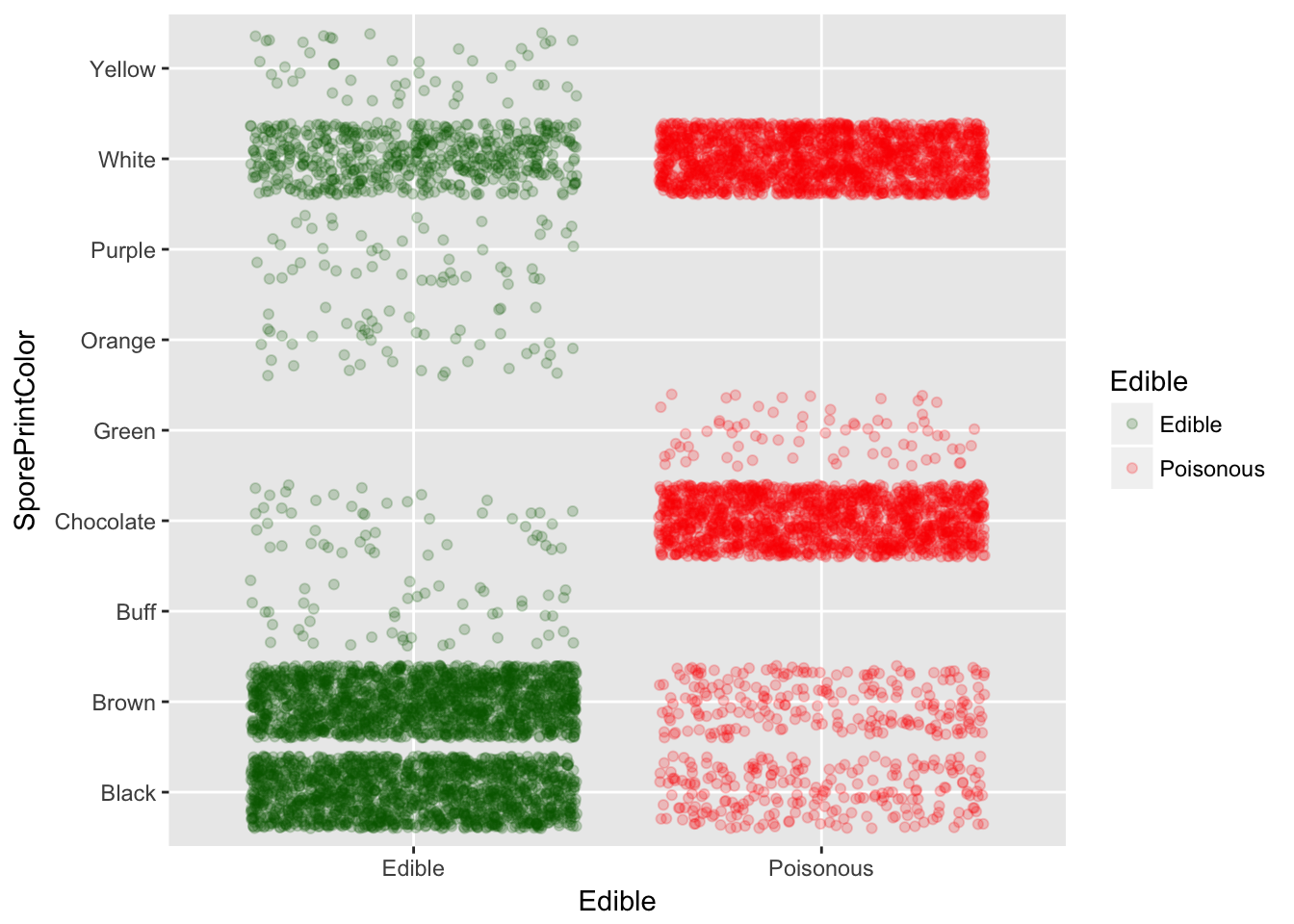
Odors fishy, foul and spicy indicate the mushroom is poisonous, so get smelling.